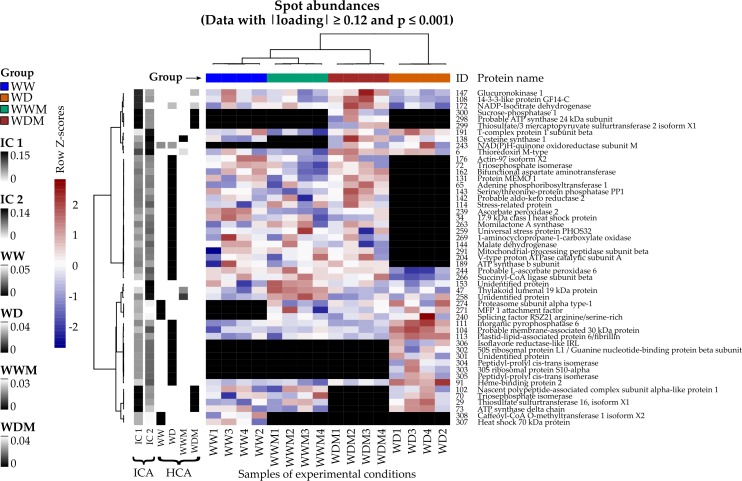
For understanding the water deficit stress mechanism in sorghum, we carried out a physiological and proteomic analysis in the leaves of Sorghum bicolor L. Moench (a drought tolerant crop mannequin) of non-colonized and colonized plants with a consortium of arbuscular mycorrhizal fungi. Physiological outcomes point out that mycorrhizal fungi affiliation enhances progress and photosynthesis in plants, under regular and water deficit situations. 2D-electrophoresis profiles revealed 51 differentially collected proteins in response to water deficit, of which HPLC/MS efficiently recognized 49.
Bioinformatics analysis of protein-protein interactions revealed the participation of totally different metabolic pathways in nonmycorrhizal in comparison with mycorrhizal sorghum plants under water deficit. In noninoculated plants, the altered proteins are associated to protein synthesis and folding (50S ribosomal protein L1, 30S ribosomal protein S10, Nascent polypeptide-associated complicated subunit alpha), coupled with a number of sign transduction pathways, guanine nucleotide-binding beta subunit (Rack1) and peptidyl-prolyl-cis-trans isomerase (ROC4).
In distinction, in mycorrhizal plants, proteins associated to vitality metabolism (ATP synthase-24kDa, ATP synthase β), carbon metabolism (malate dehydrogenase, triosephosphate isomerase, sucrose-phosphatase), oxidative phosphorylation (mitochondrial-processing peptidase) and sulfur metabolism (thiosulfate/3-mercaptopyruvate sulfurtransferase) had been discovered.
Our outcomes present a set of proteins of totally different metabolic pathways concerned in water deficit produced by sorghum plants alone or related to a consortium of arbuscular mycorrhizal fungi remoted from the tropical rain forest Los Tuxtlas Veracruz, México.
Upregulation of TDRD1 promotes the sexual maturation in allotetraploid hybridized from pink crucian carp (♀) and widespread carp (♂).
Allotetraploid Hybrids of Red Crucian Carp (Carassius auratus pink var, abbreviated as RCC) (♀) and Common Carp (Cyprinus carpio L, abbreviated as CC) (♂) is a species produced by distant hybridization. In this examine, SWATH-MS had been utilized for quantitative proteomics profiling in gonad tissue of allotetraploid and its dad and mom RCC (♀) and CC (♂) respectively.
A complete of 2338 distinctive proteins had been recognized in our proteomic profiling by SWATH-MS. Gene enrichment and network analysis based mostly on differentially expression proteins revealed some metabolic enzymes concerned in gonad progress and growth of allotetraploid.
Specially, the up-regulated gene TDRD1 (ratio=2.59, p=0.02) in allotetraploid play a big position in spermatogenesis which might pace up the sexual maturation in the course of the course of of gonad progress and growth. Protein-protein interplay and pathway analysis furtherly instructed that TDRD1 served as a hub protein in metabolic-related pathways and networks.
It might regulate gonad growth by way of regulating metabolic pathways in a synergistic approach with surrounding regulatory elements together with CS, GPIA, PGK1, IDH2. In addition, TDRD1 might instantly regulate spermatogenesis in conjunction with PIWIL1, PIWIL2 and VASA. Quantitative proteomics built-in with network analysis explored the molecular mechanism that TDRD1 regulated sexual maturation, growth and progress of allotetraploid in a synergetic approach with metabolic genes and pathways.