The Molecular INTeraction Database (MINT, ) is a public repository for protein-protein interactions (PPI) reported in peer-reviewed journals. The database grows steadily over the years and at September 2011 incorporates roughly 235,000 binary interactions captured from over 4750 publications. The net interface permits the customers to go looking, visualize and obtain interactions information.
MINT is certainly one of the members of the International Molecular Exchange consortium (IMEx) and adopts the Molecular Interaction Ontology of the Proteomics Standard Initiative (PSI-MI) requirements for curation and information alternate. MINT information are freely accessible and downloadable at http://mint.bio.uniroma2.it/mint/download.do. We report right here the development of the database, the main modifications in curation coverage and a brand new algorithm to assign a confidence to every interaction.
The Molecular INTeraction Database ( is a public repository for protein–protein interactions (PPI) reported in peer-reviewed journals. The database grows steadily over the years and at September 2011 incorporates roughly 235 000 binary interactions captured from over 4750 publications. The net interface permits the customers to go looking, visualize and obtain interactions information. MINT is certainly one of the members of the International Molecular Exchange consortium (IMEx) and adopts the Molecular Interaction Ontology of the Proteomics Standard Initiative (PSI-MI) requirements for curation and information alternate. MINT information are freely accessible and downloadable at . We report right here the development of the database, the main modifications in curation coverage and a brand new algorithm to assign a confidence to every interaction.
INTRODUCTION
Understanding the bodily and purposeful interactions between molecules in the cell is certainly one of the predominant goals of recent biology. Over the previous a long time, a number of highly effective methods have been developed to disclose, from completely different angles, the dynamics and complexity of the physiological interaction net. The retrieval, group and evaluation of those interactions are elementary to know the mobile equipment.
In the protein interaction area, a number of databases have got down to seize this data, as reported in the scientific literature, and to arrange it in a structured format in an effort to enable customers to carry out automated evaluation. However, no database has ample sources to seize and arrange all the revealed data and customers are left with the process of querying a number of databases, eager to interrogate the largest attainable dataset.
Integrating protein interaction information from completely different databases has been a problem till 2004, when the HUPO Proteomics Standards Initiative (HUPO-PSI) launched the Molecular Interaction Ontology of the Proteomics Standard Initiative (PSI-MI) XML format (present modadly accepted and applied by over 30 databases and supported by software program instruments. The detailed description of an interaction may be captured on this format, as for instance, the organic and the experimental function of the interacting proteins and the kinetic parameters.
MINT doesn’t focus on chosen mannequin organisms and in the current model incorporates interactions between proteins from greater than 30 completely different species equivalent to Homo sapiens (28 283 IE), Mus musculus (4808 IE), Rattus norvegicus (2804 IE), Drosophila melanogaster (23 534 IE), Caenorhabditis elegans (7402 IE), Saccharomyces cerevisiae (48 979 IE), Escherichia coli (4188 IE) and Helicobacter pylori (1635 IE).
NEW STRUCTURAL DIGITAL ABSTRACT
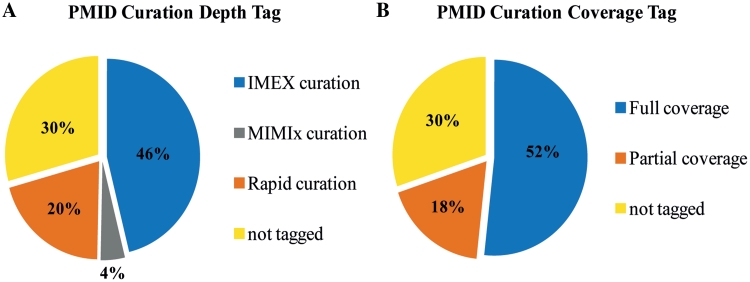
CURATION POLICY
Users can select the information that they wish to make the most of for his or her evaluation by filtering in accordance with these tags. Entries which have been curated earlier than the creation of IMEx should be unclassified whereas they’re ready to be reviewed and assigned to the appropriate class of curation customary
It is essential to level out that the entries tagged as ‘fast curation’ and/or ‘partial protection’ are in precept as correct as the IMEx entries and make the most of the identical managed vocabularies advocate by the PSI-MI consortium.
NEW FEATURES
As mentioned above, MINT incorporates entries curated to completely different annotation depth and protection.
To make this level clear in the downloadable MITAB2.6 file we now embrace a ‘curation-depth’ column that may take the values ‘IMEx’, ‘MIMIX’ or ‘fast curation’.
SCORING SYSTEM
The authentic MINT rating is predicated on a heuristic integration of the accessible proof right into a ‘mixed experimental proof’ x which is then mapped in the 0–1 interval by way of the components Score = 1−a−x.
x is computed by including up all the proof in accordance with the components where i is an index iterating over all the experimental proof, e is a coefficient that takes the worth of 1 for proof of direct interaction and 0.5 for proof that solely help and affiliation, which can be oblique, between the two companions and d displays the dimension of the experiment. Experiments are outlined giant scale if the article reporting them describes greater than 50 interactions in any other case they’re outlined small scale. This coefficient is ready to 1 for small scale and to 0.5 for big scale experiments. Finally n represents the variety of manuscripts reporting proof that help the interaction.
where i is an index iterating over all the experimental proof, e is a coefficient that takes the worth of 1 for proof of direct interaction and 0.5 for proof that solely help and affiliation, which can be oblique, between the two companions and d displays the dimension of the experiment. Experiments are outlined giant scale if the article reporting them describes greater than 50 interactions in any other case they’re outlined small scale. This coefficient is ready to 1 for small scale and to 0.5 for big scale experiments. Finally n represents the variety of manuscripts reporting proof that help the interaction.
We just lately determined to revise the scoring algorithm to appropriate some bias of the authentic algorithm and to incorporate a weight that takes under consideration ‘group recognition/belief’.
In the new model of the rating we launched the idea of built-in supporting proof y outlined as the weighted sum of the j manuscripts supporting a given interaction.
The weight of every supporting manuscript SiRi is obtained by multiplying two coefficients every various from Zero to 1 and reflecting the ‘validity’ of the experimental proof (S) or estimating the recognition/belief of the scientific group (R) respectively.
To receive S and R we first calculated s and r outlined as r = normalized citations. Where e, equally to the identical coefficient in the authentic rating, has a distinct worth in accordance with the sort of experiments supporting the interaction and emphasizes evidences of direct interaction (e = 1) with respect to experimental help that doesn’t present unequivocal proof of direct interaction, i.e. co-ip, pull down, and so forth (e = 0.5) or co-localization (e = 0.1). Conversely, r is the ratio between the variety of citations obtained by the manuscript in accordance with Google Scholar, augmented by 20 (group belief) and the variety of unbiased interactions reported in the manuscript. This latter normalization is made to consider that manuscripts describing numerous interactions have the next variety of citations, quantity that will be deceptive to make use of to measure the belief for every of the excessive variety of reported interactions.
r = normalized citations. Where e, equally to the identical coefficient in the authentic rating, has a distinct worth in accordance with the sort of experiments supporting the interaction and emphasizes evidences of direct interaction (e = 1) with respect to experimental help that doesn’t present unequivocal proof of direct interaction, i.e. co-ip, pull down, and so forth (e = 0.5) or co-localization (e = 0.1). Conversely, r is the ratio between the variety of citations obtained by the manuscript in accordance with Google Scholar, augmented by 20 (group belief) and the variety of unbiased interactions reported in the manuscript. This latter normalization is made to consider that manuscripts describing numerous interactions have the next variety of citations, quantity that will be deceptive to make use of to measure the belief for every of the excessive variety of reported interactions.
S and R are obtained by mapping to the 0—1 interval the experimental help s and the normalized variety of citations of the supporting manuscript, r. The mapping operate is of sort
 a and b are empirically set to 1.2 . Similarly to the technique utilized in the authentic MINT rating the built-in supporting proof y is mapped in the interval 0–1 in accordance with the operate
a and b are empirically set to 1.2 . Similarly to the technique utilized in the authentic MINT rating the built-in supporting proof y is mapped in the interval 0–1 in accordance with the operate