We describe a streamlined and systematic technique for cloning inexperienced fluorescent protein (GFP)-open studying body (ORF) fusions and assessing their subcellular localization in Arabidopsis thaliana cells. The sequencing of the Arabidopsis genome has made it possible to undertake genome-based approaches to find out the perform of every protein and outline its subcellular localization. This is a necessary step in direction of full practical evaluation.
The method described right here permits the economical dealing with of tons of of expressed plant proteins in a well timed vogue. We have built-in recombinational cloning of full-length trimmed ORF clones (accessible from the SSP consortium) with high-efficiency transient transformation of Arabidopsis cell cultures by a hypervirulent pressure of Agrobacterium. To reveal its utility, we have now used a variety of trimmed ORFs, representing a spread of key mobile processes and have outlined the localization patterns of 155 fusion proteins. These patterns have been categorized into 5 foremost classes, together with cytoplasmic, nuclear, nucleolar, organellar and endomembrane compartments. Several genes annotated in GenBank as unknown have been ascribed a protein localization sample.
We additionally reveal the applying of move cytometry to estimate the transformation effectivity and cell cycle section of the GFP-positive cells. This method may be prolonged to practical research, together with the exact mobile localization and the prediction of the function of unknown proteins, the affirmation of bioinformatic predictions and proteomic experiments, such because the dedication of protein interactions in vivo, and due to this fact has quite a few functions in the post-genomic evaluation of protein perform.
A gaggle of nuclear transcription elements, the Whirly proteins, had been just lately proven to be focused additionally to chloroplasts and mitochondria. In order to search out out whether or not different proteins would possibly share this function, an in silico-based screening of transcription elements from Arabidopsis and rice was carried out with the goal of figuring out putative N-terminal chloroplast and mitochondrial concentrating on sequences.
For this, the person predictions of a number of unbiased applications had been mixed to a consensus prediction using a naïve Bayes technique. This consensus prediction exhibits the next specificity at a given sensitivity worth than every of the one applications. In each species, transcription elements from a spread of protein households that possess putative N-terminal plastid or mitochondrial goal peptides in addition to nuclear localization sequences, had been discovered.
A seek for homologues inside members of the AP2/EREBP protein household revealed that focus on peptide-containing proteins are conserved amongst monocotyledonous and dicotyledonous species. Fusion of one of these proteins to GFP revealed, certainly, a twin concentrating on exercise of this protein. We suggest that dually focused transcription elements may be concerned in the communication between the nucleus and the organelles in plant cells. We additional talk about how latest outcomes on the bodily interplay between the organelles and the nucleus may have significance for the regulation of the localization of these proteins.
Arabidopsis thaliana histone deacetylase 1 (AtHD1 or AtHDA19), a homolog of yeast RPD3, is a world regulator of many physiological and developmental processes in vegetation. In spite of the genetic proof for a task of AtHD1 in plant gene regulation and improvement, the biochemical and mobile properties of AtHD1 are poorly understood. Here we report mobile localization patterns of AtHD1 in vivo and histone deacetylase exercise in vitro.
The transient and steady expression of a inexperienced fluorescent protein (GFP)-tagged AtHD1 in onion cells and in roots, seeds and leaves of the transgenic Arabidopsis, respectively, revealed that AtHD1 is localized in the nucleus presumably in the euchromatic areas and excluded from the nucleolus. The localization patterns of AtHD1 are completely different from these of AtHD2 and AtHDA6 which are concerned in nucleolus formation and silencing of transgenes and repeated DNA parts, respectively.
In addition, a histone deacetylase exercise assay confirmed that the recombinant AtHD1 produced in micro organism demonstrated a particular histone deacetylase exercise in vitro. The information recommend that AtHD1 is a nuclear protein and possesses histone deacetylase actions accountable for world transcriptional regulation essential to plant progress and improvement.
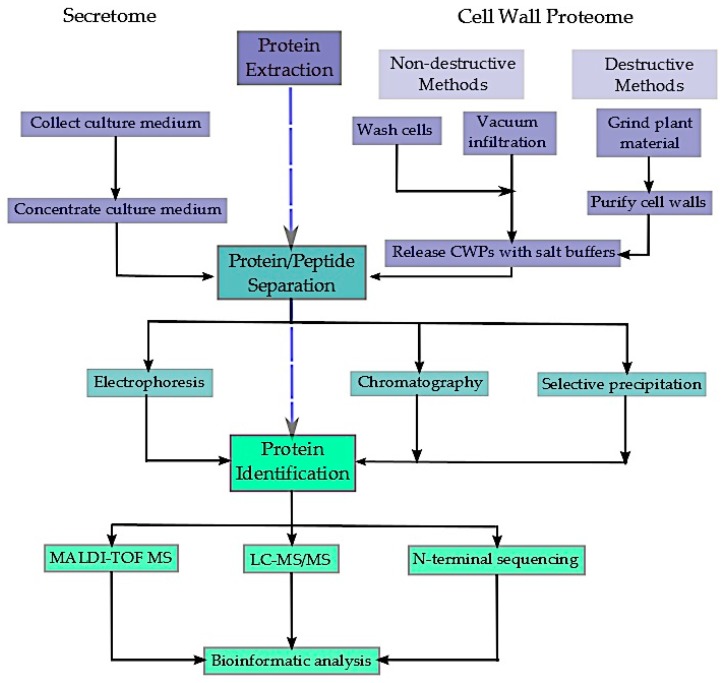
Proteins secreted by plant cells into the extracellular area, consisting of the cell wall, apoplastic fluid, and rhizosphere, play essential roles throughout improvement, nutrient acquisition, and stress acclimation. However, isolating the total vary of secreted proteins has confirmed troublesome, and new methods are continuously evolving to extend the quantity of proteins that may be detected and recognized. In addition, the dynamic nature of the extracellular proteome presents the additional problem of figuring out and characterizing the post-translational modifications (PTMs) of secreted proteins, significantly glycosylation and phosphorylation.
Such PTMs are frequent and essential regulatory modifications of proteins, taking part in a key function in many organic processes. This evaluate explores the newest strategies in isolating and characterizing the plant extracellular proteome with a deal with the mannequin plant Arabidopsis thaliana, highlighting the present challenges but to be overcome. Moreover, the essential function of protein PTMs in cell wall signalling, improvement, and plant responses to biotic and abiotic stress is mentioned.
The expression of auxin-responsive genes is regulated by the TIR1/AFB auxin receptor-dependent degradation of Aux/IAA transcriptional repressors, which work together with auxin-responsive elements (ARFs). Most of the 29 Aux/IAA genes current in Arabidopsis haven’t been functionally characterised thus far. IAA8 seems to have a definite perform from the opposite Aux/IAA genes, attributable to its distinctive transcriptional response to auxin and the soundness of its encoded protein.
In this examine, we characterised the perform of Arabidopsis IAA8 in varied developmental processes ruled by auxin and in the transcriptional regulation of the auxin response. Transgenic vegetation expressing estrogen-inducible IAA8 (XVE::IAA8) exhibited considerably fewer lateral roots than the wild kind, and an IAA8 loss-of-function mutant exhibited considerably extra. Ectopic overexpression of IAA8 resulted in irregular gravitropism.
The robust induction of early auxin-responsive marker genes by auxin therapy was delayed by IAA8 overexpression. GFP-fusion evaluation revealed that IAA8 localized not solely to the nucleus, however, in distinction to different Aux/IAAs, additionally to the cytosol. Furthermore, we demonstrated that IAA8 interacts with TIR1, in an auxin-dependent vogue, and with ARF proteins, each in yeast and in planta. Taken collectively, our outcomes present that IAA8 is concerned in lateral root formation, and that this course of is regulated by means of the interplay with the TIR1 auxin receptor and ARF transcription elements in the nucleus.
Dynamic conduct of Arabidopsis eIF4A-III, putative core protein of exon junction advanced: quick relocation to nucleolus and splicing speckles beneath hypoxia
Here, we establish the Arabidopsis thaliana ortholog of the mammalian DEAD field helicase, eIF4A-III, the putative anchor protein of exon junction advanced (EJC) on mRNA. Arabidopsis eIF4A-III interacts with an ortholog of the core EJC part, ALY/Ref, and colocalizes with different EJC parts, resembling Mago, Y14, and RNPS1, suggesting an identical perform in EJC meeting to animal eIF4A-III.
A inexperienced fluorescent protein (GFP)-eIF4A-III fusion protein confirmed localization to a number of subnuclear domains: to the nucleoplasm throughout regular progress and to the nucleolus and splicing speckles in response to hypoxia. Treatment with the respiratory inhibitor sodium azide produced an similar response to the hypoxia stress. Treatment with the proteasome inhibitor MG132 led to accumulation of GFP-eIF4A-III primarily in the nucleolus, suggesting that transition of eIF4A-III between subnuclear domains and/or accumulation in nuclear speckles is managed by proteolysis-labile elements.
As revealed by fluorescence restoration after photobleaching evaluation, the nucleoplasmic fraction was extremely cellular, whereas the speckles had been the least cellular fractions, and the nucleolar fraction had an intermediate mobility. Sequestration of eIF4A-III into nuclear swimming pools with completely different mobility is more likely to mirror the transcriptional and mRNA processing state of the cell.