Fresh fish are extremely perishable meals merchandise and their brief shelf-life limits their industrial exploitation and results in waste, which has a detrimental influence on aquaculture sustainability. New non-thermal meals processing strategies, resembling high pressure (HP) processing, extend shelf-life whereas assuring high meals quality.
The impact of HP processing (600MPa, 25 °C, 5min) on European sea bass (Dicentrarchus labrax) fillet quality and shelf life was investigated. The data offered includes microbiome and proteome profiles of management and HP-processed sea bass fillets from 1 to 67 days of isothermal storage at 2 °C.
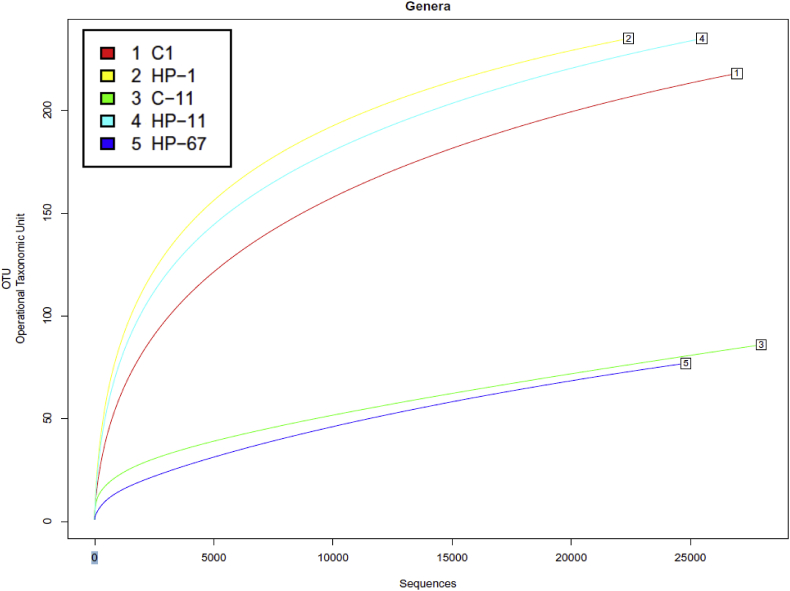
Bacterial variety was analysed by Illumina high-throughput sequencing of the 16S rRNA gene in pooled DNAs from management or HP-processed fillets after 1, 11 or 67 days and the uncooked reads had been deposited within the NCBI-SRA database with accession quantity PRJNA517618.
Yeast and fungi variety had been analysed by high-throughput sequencing of the interior transcribed spacer (ITS) area for management and HP-processed fillets on the finish of storage (11 or 67 days, respectively) and have the SRA accession quantity PRJNA517779. Quantitative label-free proteomics profiles had been analysed by SWATH-MS (Sequential Windowed data unbiased Acquisition of the Total High-resolution-Mass Spectra) in myofibrillar or sarcoplasmic enriched protein extracts pooled for management or HP-processed fillets after 1, 11 and 67 days of storage. Proteome data was deposited within the ProteomeXchange Consortium through the PRIDE associate repository with the dataset identifiers PXD012737.
Connecting Histopathology Imaging and Proteomics in Kidney Cancer by means of Machine Learning.
Proteomics data encode molecular options of diagnostic worth and precisely mirror key underlying organic mechanisms in cancers. Histopathology imaging is a well-established medical method to most cancers prognosis.
The predictive relationship between large-scale proteomics and H&E-stained histopathology pictures stays largely uncharacterized. Here we examine such associations by means of the appliance of machine studying, together with deep neural networks, to proteomics and histology imaging datasets generated by the Clinical Proteomic Tumor Analysis Consortium (CPTAC) from clear cell renal cell carcinoma sufferers.
We report sturdy correlations between a set of diagnostic proteins and predictions generated by an imaging-based classification mannequin. Proteins considerably correlated with the histology-based predictions are considerably implicated in immune responses, extracellular matrix reorganization, and metabolism.
Moreover, we confirmed that the genes encoding these proteins additionally reliably recapitulate the organic associations with imaging-derived predictions based mostly on robust gene-protein expression correlations. Our findings provide novel insights into the integrative modeling of histology and omics data by means of machine studying, in addition to the methodological foundation for brand spanking new analysis alternatives on this and different most cancers varieties.