The PRoteomics IDEntifications (PRIDE) database is one of the world-leading information repositories of mass spectrometry (MS)-based proteomics information. Since the starting of 2014, PRIDE Archive (http://www.ebi.ac.uk/pride/archive/) is the new PRIDE archival system, changing the authentic PRIDE database.
Here we summarize the developments in PRIDE sources and related tools since the earlier update manuscript in the Database Issue in 2013. PRIDE Archive constitutes a whole redevelopment of the authentic PRIDE, comprising a brand new storage backend, information submission system and net interface, amongst different elements.
PRIDE Archive helps the most-widely used PSI (Proteomics Standards Initiative) information normal codecs (mzML and mzIdentML) and implements the information necessities and tips of the ProteomeXchange Consortium. The extensive adoption of ProteomeXchange inside the group has triggered an unprecedented improve in the quantity of submitted information units (round 150 information units per thirty days).
We define some statistics on the present PRIDE Archive information contents. We additionally report on the standing of the PRIDE related stand-alone tools: PRIDE Inspector, PRIDE Converter 2 and the ProteomeXchange submission device. Finally, we are going to give a quick update on the sources beneath growth ‘PRIDE Cluster’ and ‘PRIDE Proteomes’, which offer a complementary view and quality-scored info of the peptide and protein identification information obtainable in PRIDE Archive.
The PRoteomics IDEntifications (PRIDE) database is one of the world-leading information repositories of mass spectrometry (MS)-based proteomics information. Since the starting of 2014, PRIDE Archive is the new PRIDE archival system, changing the authentic PRIDE database. Here we summarize the developments in PRIDE sources and related tools since the earlier update manuscript in the Database Issue in 2013.
PRIDE Archive constitutes a whole redevelopment of the authentic PRIDE, comprising a brand new storage backend, information submission system and net interface, amongst different elements. PRIDE Archive helps the most-widely used PSI (Proteomics Standards Initiative) information normal codecs (mzML and mzIdentML) and implements the information necessities and tips of the ProteomeXchange Consortium.
The extensive adoption of ProteomeXchange inside the group has triggered an unprecedented improve in the quantity of submitted information units (round 150 information units per thirty days). We define some statistics on the present PRIDE Archive information contents. We additionally report on the standing of the PRIDE related stand-alone tools: PRIDE Inspector, PRIDE Converter 2 and the ProteomeXchange submission device. Finally, we are going to give a quick update on the sources beneath growth ‘PRIDE Cluster’ and ‘PRIDE Proteomes’, which offer a complementary view and quality-scored info of the peptide and protein identification information obtainable in PRIDE Archive.
INTRODUCTION
Data sharing and public availability of proteomics information requires substantial infrastructure. Several public repositories and sources have been developed in the final decade, every with totally different goals in thoughts. Among them, the PRoteomics IDEntifications (PRIDE) database was initially established at the European Bioinformatics Institute (EBI) in 2004 (1) to allow public information deposition of mass spectrometry (MS) proteomics information, offering entry to the experimental information described in scientific publications. Since then, PRIDE has been evolving to meet the ever-growing necessities of the proteomics group (2–5). Since January 2014, PRIDE Archive (http://www.ebi.ac.uk/pride/archive/) is the new PRIDE archival useful resource, changing the authentic PRIDE database (1).
Among different information varieties, PRIDE Archive can retailer peptide and protein identifications and related quantification values, the corresponding mass spectra (each as processed peak lists and uncooked information), gel pictures, the searched sequence databases or spectral libraries, programming scripts and some other technical and/or organic metadata supplied by the submitters. It is vital to focus on that PRIDE Archive shops all processed outcomes as initially analyzed by the authors.
In this manuscript, we are going to summarize the PRIDE related developments of the final three years, since the earlier Nucleic Acids Research (NAR) database update manuscript was printed (). We will describe the new PRIDE Archive in greater element however can even present details about the standalone tools and define the ongoing high quality management (QC) efforts, and lastly focus on future developments.
MAIN CHARACTERISTICS OF PRIDE ARCHIVE
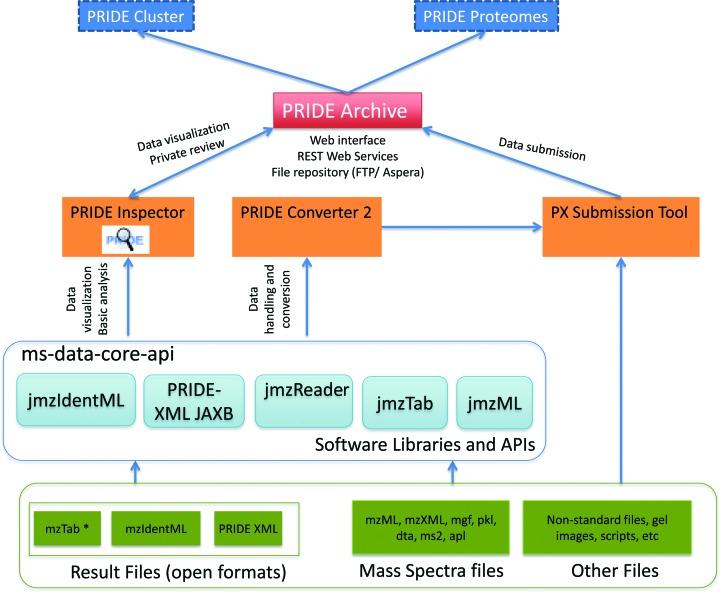
As talked about above, PRIDE Archive fully changed the authentic PRIDE database system since the starting of 2014. Figure offers an outline of the present tools, sources and software program included in the PRIDE ecosystem.
PRIDE Archive fully depends on information set submissions made by researchers. Original submitted information are usually not modified, however PRIDE curators can add some further metadata annotations at the degree of the information set (e.g. tagging of information units or the printed reference/s). Data units stay non-public (password protected) by default and are solely made publicly obtainable after the related manuscript has been accepted, or when the PRIDE crew is notified to take action by the authentic submitter.
Types of information units and help for open normal codecs
PRIDE Archive is an information set or challenge centric useful resource. An information set consists of a number of experimental assays. This method has advanced from the authentic PRIDE system, the place information had been organised per experiment (equal to an ‘assay’), usually corresponding to at least one MS run. Following the frequent identifier house applied inside the PX Consortium, all PRIDE Archive information units are actually named utilizing PXD identifiers
There are two varieties of information units that may be submitted, based mostly on the PX tips: ‘Complete’ and ‘Partial’. For each varieties, a set of frequent experimental metadata (agreed by all the PX companions) and uncooked information mass spectra are all the time obtainable. The distinction between each varieties is related to the manner the processed identification outcomes are supplied.
In the case of a ‘Complete’ information set it’s attainable for PRIDE to instantly join the processed peptide/protein identification outcomes with the submitted mass spectra. This can solely be achieved if the processed outcomes can be found in a supported open information format for peptide/protein identification information. PRIDE Archive totally helps PRIDE XML (the authentic PRIDE information format) and the PSI (Proteomics Standards Initiative) mzIdentML information normal format for identification information
For the latter, the accompanying mass spectra recordsdata should even be submitted since mzIdentML doesn’t comprise precise mass spectra information. ‘Complete’ information units may be break up in assays (corresponding to every particular person PRIDE XML or mzIdentML file) and as such, they are often annotated in greater element by submitters. Assays get a secondary accession quantity as properly (an integer quantity), equal to the identifiers given to PRIDE experiments in the authentic PRIDE system. All ‘Complete’ information units additionally get a Digital Object Identifier (DOI) to enhance the monitoring of information units and enhance recognition for submitters.
In the case of ‘Partial’ information units , the spectra and the identification outcomes can’t be related in a simple manner since the processed outcomes are usually not obtainable in an open format supported by PRIDE. Instead, the corresponding evaluation software program output recordsdata (in heterogeneous codecs) are made obtainable. In this case, assay numbers and DOIs are usually not assigned and the annotated experimental metadata is relevant to the information set as a complete. Thanks to this mechanism, information units generated utilizing any proteomics experimental method (other than information units containing solely SRM information, that are saved in the PX PASSEL useful resource) can now be saved in PRIDE Archive. This consists of for example top-down proteomics, information unbiased acquisition approaches (similar to the SWATH-MS, MSe and HD-MSe methods, amongst others) and MS imaging information units. For the latter sort of information units, a extra formal submission mechanism has been developed, involving the necessary presence of sure MS imaging information varieties and recordsdata
Finally, the information units obtainable in the authentic PRIDE database earlier than PX had been assigned a brand new ‘PRD’ identifier, along with the authentic PRIDE experiment numbers. For these older information units, uncooked information are usually not obtainable though peak lists can be found typically.
Technical Implementation, together with the inside file submission pipeline
All PRIDE related software program is open supply and written in Java. PRIDE Archive supply code is obtainable in GitHub The software program structure of PRIDE Archive has two major elements: (i) the PRIDE Archive net and net service for presenting and looking the information units and (ii) the inside submission pipeline for dealing with incoming submissions and retailer them. Detailed technical info (together with figures) is obtainable as Supplementary File 1.
The design of the net and net service follows the ‘Layered Architecture Pattern’ the place every layer serves just one major accountability and can solely entry the layer beneath (see Supplementary File 1). One of major challenges in designing similar to a system is to give you a scalable answer for storing and looking giant quantities of information. We adopted a polyglot persistent mannequin the place the conventional relational database and the new NoSQL information shops are combined to ship efficiency. An Oracle database) is used as the relational retailer for the organic and technical metadata, and a number of Solr share used for storing and enabling the search performance for protein and peptide identifications, and spectra.
The PRIDE Archive inside submission pipeline has three levels (Fig (i) file validation, the place schema compliance is ensured for the XML-based file codecs. In addition, a report is generated that’s checked by PRIDE curators to make sure that the information are right and may be submitted (ii) information submission, the place the information are literally submitted to PRIDE Archive.
As a consequence, the submitter will get the information set PXD accession quantity, DOI (in the case of ‘Complete’ information units) and the reviewer credentials for accessing the information privately and (iii) information publication, to make the information publicly obtainable as soon as it’s applicable. In addition, some inside file conversion takes place (see beneath) and the details about the new information units will get transmitted to the PX portal ProteomeCentral.